A team involving Kobe University researchers has succeeded in developing ‘Target-AID’, a genome editing technique that does not cleave the DNA. The technique offers, through high-level editing operation, a method to address the existing issues of genome editing. It is expected that the technique will be applied to gene therapy in the future in addition to providing a powerful tool for breeding useful organisms and conducting disease and drug-discovery research. The findings were published online in Science on August 5 (Japan Standard Time).
The team consists of Project Associate Professor NISHIDA Keiji and Professor KONDO Akihiko (Graduate School of Science, Technology and Innovation, Kobe University) as well as Associate Professor YACHIE Nozomu (Synthetic Biology Division, Research Center for Advanced Science and Technology, the University of Tokyo) and Professor HARA Kiyotaka (Department of Environmental Sciences, Graduate School of Nutritional and Environmental Sciences, University of Shizuoka).
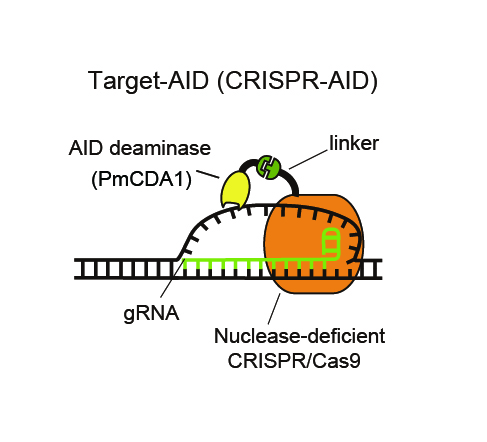
Deaminase is attached by a linker to nuclease –deficient CRISPR/Cas9. Guide RNA recognizes the DNA sequence of target genome and the deaminase modifies the base of the unwound DNA.
Genome editing - which can directly manipulate the genome information of various organisms without leaving an artificial strand - has seen rapid progress in recent years and it is gradually becoming a revolutionary tool in fields ranging from life sciences to advanced medical research.
Known to be highly effective, genome editing using ‘artificial nuclease’ (1) aims to cut the DNA at the target point and to modify the gene while it is repaired. As this method is efficacious in biological materials for which genetic operation was difficult, it has been used mainly for higher eukaryotes such as animals and plants.
However, a major issue with this method has been the uncertainty of inducing the intended mutation and ‘cytotoxicity’ (2) caused by splitting of chromosomes.
In the present research, researchers demonstrated that it is possible to modify genetic function by inducing target ‘point mutation’ (3) at a highly efficient rate (Figure 2). The point mutation was induced by forming a synthetic complex (Figure 1) through removal of nuclease activity from the CRISPR system – a technique using artificial nuclease – and addition of deaminase, a deaminizing (base-modifying) enzyme, and then expressing it in yeasts and mammalian cells. Moreover, it was confirmed that, compared to previous nuclease models, cytotoxicity is significantly reduced by modifying the DNA without cutting.
The present technique can realize a more diverse, higher-level genome editing operation as it can efficiently conduct intended modification in a way that does not put significant burden on the cells. It is expected that the new technique will be applied to gene therapy in the future in addition to providing a powerful tool for breeding useful organisms and conducting disease and drug-discovery research.
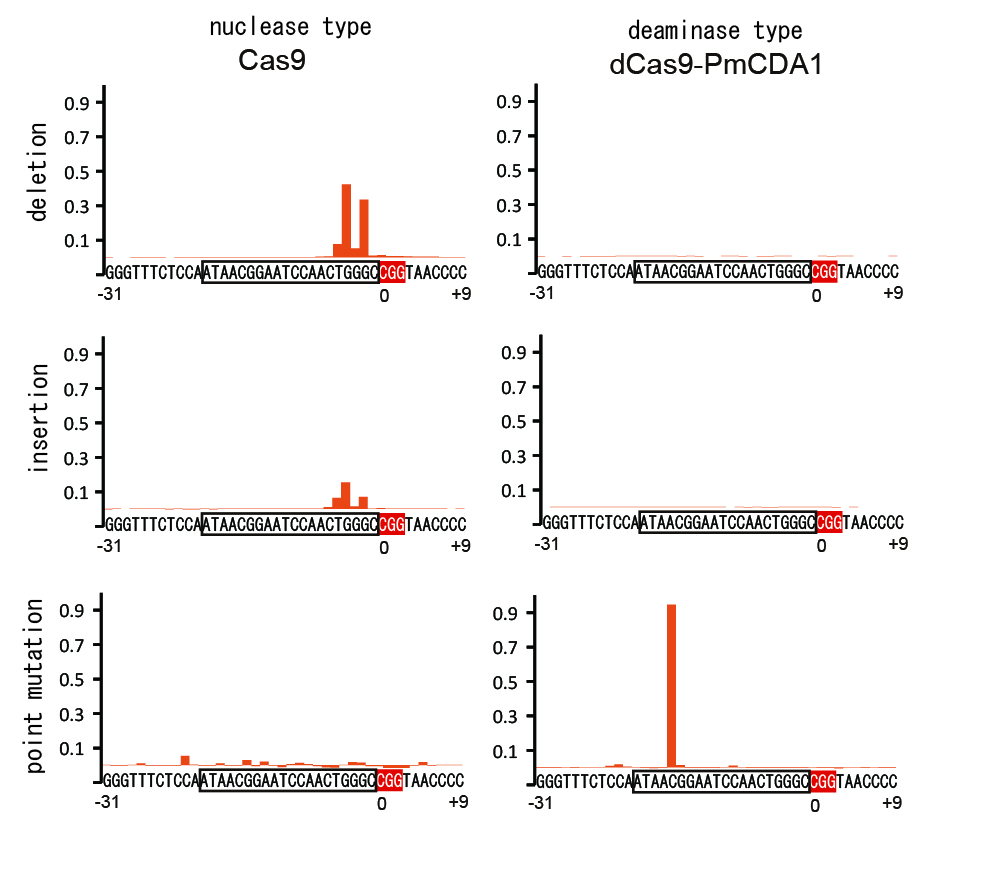
The figure analyzes the frequency at which mutation was induced in 40 bases of the target DNA sequence. In the nuclease model (left) mainly insertion and deletion are induced but in the deaminase model only point mutation is induced.
Technical terms
- 1. Artificial nuclease:
- An enzyme artificially designed for recognizing and cutting arbitrary sequence of DNA strand.
- 2. Cytotoxicity:
- The quality of causing cell death or functional disorder within cells.
- 3. Point Mutation:
- A sudden mutation in which one base is changed into another base.
Acknowledgements
- Ministry of Education, Culture Sports and Technology (MEXT) Creation of Innovative Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe);
- MEXT Special Coordination Fund for Promoting Science and Technology - Ministry of Economy, Trade and Industry (METI) Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterial
- Cross-ministerial Strategic Innovation Promotion Program (SIP) ‘Technologies for creating next-generation agriculture, forestry and fisheries’
Journal information
- Title
- “ Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems ”
- DOI
- 10.1126/science.aaf8729
- Authors
- Keiji Nishida, Takayuki Arazoe, Nozomu Yachie, Satomi Banno, Mika Kakimoto, Mayura Tabata, Masao Mochizuki, Aya Miyabe, Michihiro Araki, Kiyotaka Y. Hara, Zenpei Shimatani and Akihiko Kondo
- Journal
- Science








