Researchers have shed light on the distribution of Japanese eel by analyzing environmental DNA (eDNA) from small samples of river water. This could enable faster and more effective surveys of Japanese eel populations, and help to conserve this endangered species. The finding was published on February 27 in Aquatic Conservation: Marine and Freshwater Ecosystems.


Eels are migratory fish that spawn in the ocean and grow up along the coast and in rivers. There are 16 known species in the world, distributed in 150 countries. The Japanese eel (Anguilla japonica) is found across East Asia. Since ancient times it has been an important part of Japanese life: as a food source, a subject of traditional poems and art, and sometimes even as a target of worship. However, eel catches have fallen drastically since the 1970s, and in 2014 it was added to the International Union for Conservation of Nature (IUCN) Red List of Threatened Species.
Most river surveys of Japanese eel use electrofishing. However, this method requires a lot of time and resources, and for widely distributed species it may not collect enough data. Surveys are usually carried out in the daytime, while the nocturnal eels hide among vegetation and dirt.
Rapidly-advancing eDNA technology can monitor aquatic lifeforms through extraction and analysis of DNA present in water, without capturing the organisms themselves. In this study, the team investigated whether eDNA analysis could be used to show the distribution of Japanese eel. They collected 1-liter samples from 125 locations upstream and downstream in 10 rivers in Japan, and analyzed the eDNA from these samples using a Real-Time PCR system. At the same time they carried out an electrofishing survey in the same locations, and compared this with the eDNA analysis results.

sampling investigation using electrofishing (left), water sample for eDNA analysis (right)
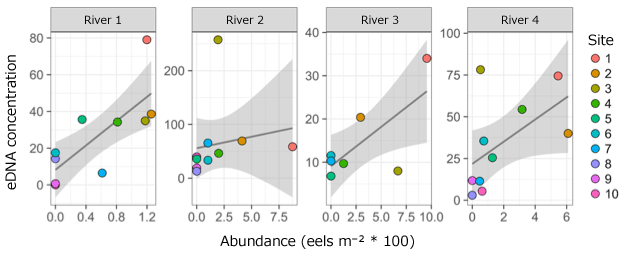
Japanese eel eDNA was found in 91.8% of the locations where eel had been confirmed using electrofishing (56 of 61 locations), and eDNA was also detected in an additional 35 areas (mainly upstream) where eel individuals were not found. This shows that eDNA analysis is more sensitive than conventional surveys for detecting the presence of Japanese eel in rivers. Electrofishing data for eel numbers and biomass also positively correlated with eDNA concentrations, showing that eDNA could help us estimate the abundance and biomass of Japanese eel.
In this study, electrofishing required three or more people for each river and took at least three days. Collecting water samples for eDNA analysis only needed two people, took half a day at the most, and data processing was finished by one person in one and a half days. When carrying out a large-scale distribution survey the eDNA analysis method is better in terms of human and time resources.
This method could potentially survey populations on an even wider scale. It is non-lethal, making it ideal for monitoring endangered species. The team is currently using eDNA analysis to monitor eels in Japan and overseas: it can be used as an international unified method for widely-distributed species. This could be a great help in the conservation and sustainable use of eel species worldwide.
The eDNA analysis method is also effective in dealing with the invasion of foreign eel species. For 20 years there have been reports of foreign eels (European eels and American eels) being released into Japanese waterways. These species look the same as Japanese eel, making them hard to detect. They are also long-lived so they may impact the ecosystem over long periods of time. By carrying out a wide-ranging investigation using eDNA analysis, we can swiftly identify foreign eel species and their distribution.
This study was carried out by Research Associate Hikaru Itakura (Kobe University Graduate School of Science), Assistant Professor Ryoshiro Wakiya (Chuo University), Assistant Professor Satoshi Yamamoto (Kyoto University), Associate Professor Kenzo Kaifu (Chuo University), Associate Professor Takuya Sato and Associate Professor Toshifumi Minamoto (both from Kobe University).
Itakura comments: “Concentration of eDNA in rivers is influenced by physical properties such as water depth and the speed of the current. Next we must increase the accuracy of eDNA analysis by clarifying the impact of these physical properties on eDNA concentration.”
Journal information
- Title
- “Environmental DNA analysis reveals the spatial distribution, abundance and biomass of Japanese eels at the river basin scale”
- DOI
- 10.1002/aqc.3058
- Authors
- Research Associate Hikaru Itakura (Kobe University)
Assistant Professor Ryoshiro Wakiya (Chuo University)
Assistant Professor Satoshi Yamamoto (Kyoto University)
Associate Professor Kenzo Kaifu (Chuo University)
Associate Professor Takuya Sato (Kobe University)
Associate Professor Toshifumi Minamoto (Kobe University) - Journal
- Aquatic Conservation: Marine and Freshwater Ecosystems