A new method to assess the status of immune responses to specific antigens in detail by analysis using the B cell receptor (BCR) repertoire(*1) has been developed by a research group, consisting of Assistant Professor Yohei Funakoshi, Associate Professor Kimikazu Yakushijin, Professor Hironobu Minami (Kobe University Graduate School of Medicine), Associate Professor Goh Ohji (Kobe University Hospital), and Researcher Takaji Matsutani (Repertoire Genesis Inc.).
These research results were published online in British Journal of Haematology on June 22, 2023.
Main Points
- Previously, B-cell receptor (BCR) repertoire analysis has provided a huge amount of gene sequence data of antibodies (B cell receptors) , but it was difficult to use this data to evaluate the immunological reactions to specific antigens.
- While conducting the repertoire analysis, it was found that the immunological reactions could be evaluated by “sampling over time in the short time period wherein immunity is activated” and “utilizing the BCR gene sequence database for specific antigens”.
- This analysis is not the conventional analysis using protein level such as “the measurement of antibody titers using the ELISA method”, but analysis using mRNA level, and so evaluation is possible for samples with extremely elevated antibody titers after prophylactic administration of antibody preparations (proteins).
- Detailed information can be derived, such as antibody epitopes expressed in immunological reactions or neutralization activity and the like, if there is additional information in respect of the antibodies in the databases used.
- By utilizing this analysis method to investigate immune responses following the administration of mRNA vaccines (mRNA formulations), it will be possible to widely evaluate the functionality of mRNA formulations.
Background of the Research
In recent years, genetically engineered antibody preparations (tixagevimab/cilgavimab*2) have been widely administered as preventative drugs, primarily for cases with compromised immune systems where there is insufficient antibody production after inoculation with mRNA SARS-CoV-2 vaccines. However, antibody prophylaxis is not a complete substitute for vaccination, and it is recommended that regular vaccines be administered even in cases where prophylaxis is administered. In such cases, the immunological reaction is typically assessed after vaccine inoculation by measuring antibodies (proteins) using the ELISA method. However, due to the presence of significant amounts of antibodies from the prophylactic preparations, the reactions after vaccine inoculation cannot be accurately evaluated using the ELISA method. For these reasons, there is a need for an assessment method to evaluate the immune response after vaccine inoculation in these cases, focusing on mRNA levels rather than protein levels.
The research group considers that BCR repertoire analysis, which analyses the sequences of antibodies (BCR) using mRNA, is an effective method for mRNA level evaluation. However, there was no well-established assessment method for specific antigen reactions utilizing huge repertoire data analysis.
Content of the Research
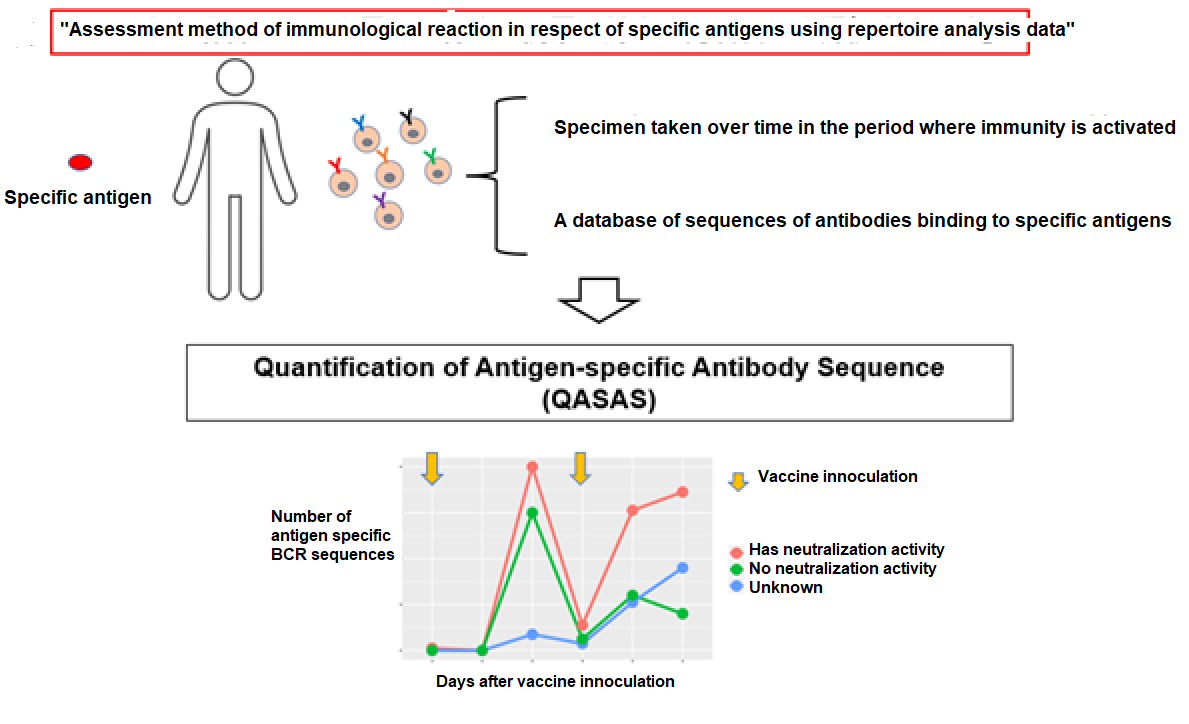
In this research, by conducting “sampling over time in the short time period wherein immunity is activated” and “availing of the BCR gene sequence database in respect of specific antigens” in implementing repertoire analysis, the researchers found that it was possible to assess the immunological reactions in respect of specific antigens at the mRNA level (FIG. 1). Specifically, it is considered that the initial immunity is activated at about two weeks after inoculation with mRNA SARS CoV-2 vaccine, and the researchers performed repertoire analysis at about one week after inoculation when additional immunity is considered to be activated (FIG. 1). On this occasion, the sequence information derived by repertoire analysis is extremely large, and it is difficult to assess the immunological reaction in respect of specific antigens using repertoire analysis alone. For that reason, against the background of the recent COVID-19 pandemic, the researchers focused on the fact that a vast amount of information on SARS-CoV-2 antibody sequences has been accumulated and compiled into databases. Using this database, they focused on sequences that were reported to bind to a specific antigen (spike protein) , or sequences similar to it, and conducted analysis (Figure 1), enabling them to evaluate post-vaccination responses (Figure 2).
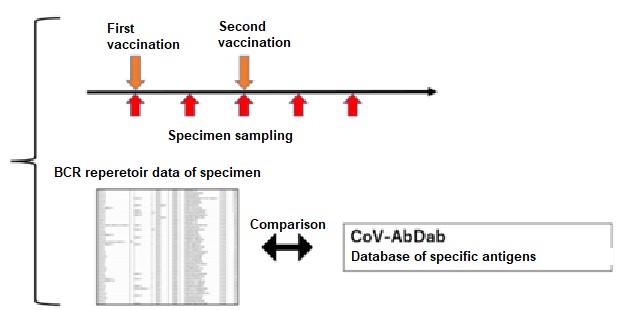
This procedure avails of "B cell receptor (BCR) repertoire data relating to the activation period of the humoral immunity" and "the database of the BCR sequences binding to the target antigen". The group analyzed how many of all BCR sequences obtained by BCR Repertoire analysis contain antigen-specific BCR sequences in the database., Credit: Yohei Funakoshi, License:CC BY
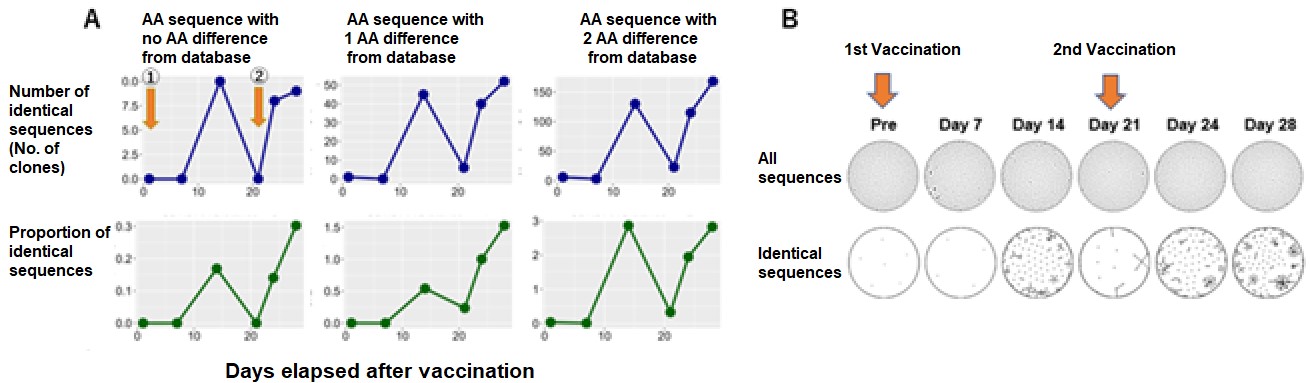
The overtime variations in the number, frequency and clusters of matched SARS-CoV-2 specific sequences after the initial and second vaccine inoculation. Healthy volunteers received the first dose (1) of mRNA SARS-CoV-2 vaccine (monovalent BNT162b2) and the second dose (2) 21 days later. The researchers searched for SARS-CoV-2 specific sequences from the BCR repertoire data after these vaccine inoculations. Modified from journal article., Credit: © 2023 British Society for Haematology and John Wiley & Sons Ltd., Restriction:Permission from journal required.
Because this method analyses post-vaccine response at the mRNA level, it can be used to evaluate post-vaccine response in immunocompromised patients who have received antibody prophylaxis. In practice, patients who received recombinant antibodies (tixagevimab/silgavimab) after hematopoietic stem cell transplantation have shown extremely high antibody titers as assessed by ELISA after administration of antibody drugs (the black line in Figure 3). Even with such a large amount of antibody protein in the body, the research group’s method was able to clearly demonstrate a post-vaccination response (the blue line in Figure 3).
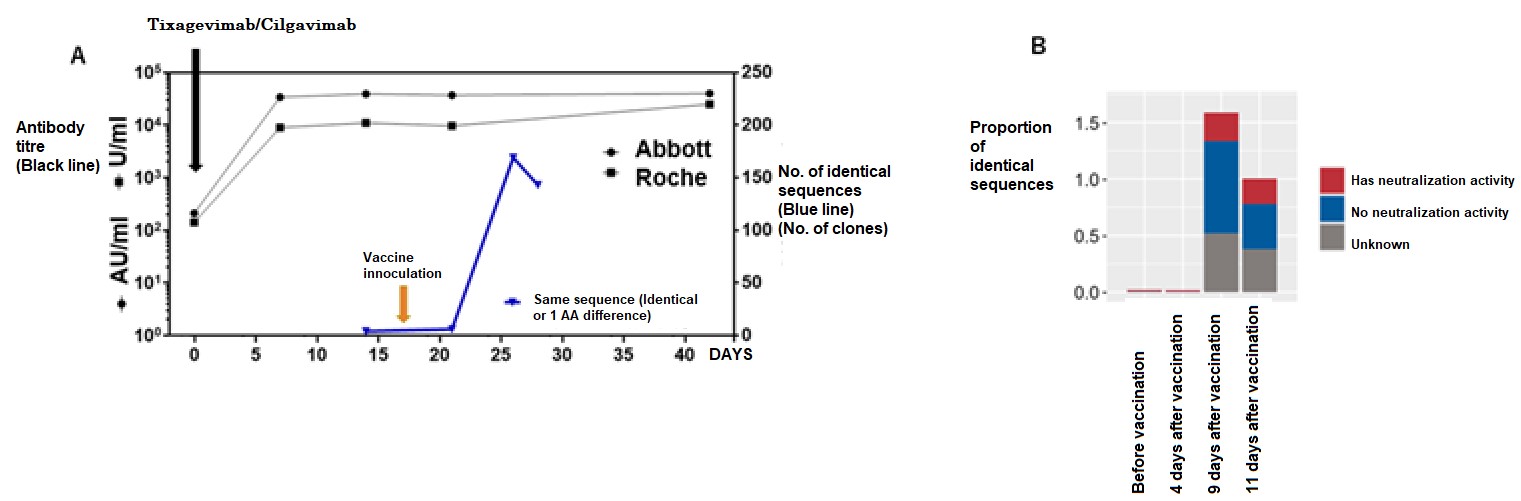
A: Tixagevimab/Cilgavimab was administered to cases after umbilical cord blood transplant and were inoculated with the mRNA vaccine thereafter (yellow arrow). The anti-SARS-CoV-2 antibody titer (black line) transitioned to an extremely high titer value after prophylactic antibody administration, but even then the researchers were able to assess the reactions after vaccine inoculation by assessing SARS-CoV-2 specific sequences (Blue line). B: By availing of the neutralization activity data of antibodies in the database, the research group was able to assess the characteristics of the elevated antibodies after vaccine inoculation. Modified from journal article., Credit: © 2023 British Society for Haematology and John Wiley & Sons Ltd., Restriction:Permission from journal required.
Further Developments
The research group termed the newly developed “Assessment method of immunological reaction in respect of specific antigens using repertoire analysis data (patent application filed)" as the "Quantification of antigen specific antibody sequence (QASAS)" method (Figure 4). The method can assess immune response even in situations where large amounts of antibodies are present in the body, e.g. due to administration of antibody drugs or infection/vaccination. In addition, the advantages of this method don't stop there. If there is epitope or neutralization activity information in the database used, the assessment of the properties of the antibodies created from the immunologically reactive BCR sequence is enabled (Figure. 3B). The QASAS method has various advantages that the conventional ELISA method lacks, and it is considered suitable for assessing a wide range of mRNA formulations that are still under development.
Credit: Yohei Funakoshi, License:CC BY
Explanation of terminology
- *1 Repertoire analysis of B cell receptors
- B cells which are the main lymphocytes express B cell receptors (BCR) which are receptor molecules recognizing antigens. When these receptors bind to antigens such as those of cancer or viruses, the cells are activated, initiating an immune reaction. To be able to react with diverse antigens, diverse BCRs are created by mechanisms of genetic rearrangement and somatic hypermutation. The extent of this diversity is estimated to be as high as 10 to the 15th power in BCRs. This collection of lymphocytes characterized by BCRs with individually distinct specificities is called a BCR repertoire. Repertoire is a French word synonymous with repertory.
- *2 Tixagevimab/Cilgavimab
- Anti-SARS-CoV-2 monoclonal antibodies indicated for the suppression of infection and pathogenesis caused by ARS-CoV-2, consisting of two long-acting antibodies, tixagevimab and silgavimab, derived from B cells provided by patients who have recovered from SARS-CoV-2 infection.
- *3 CoV-AbDab
- CoV-AbDab is a database managed by Oxford Protein Informatics Group (Department of Statistics, University of Oxford) and includes data relating to disclosed antibodies and patented antibodies which are known to bind to at least one [type of] beta corona virus, and nano bodies.
Acknowledgments
The COVID-19 infection status of the cases in this study (confirmation of uninfected status) was evaluated by ELISA against SARS-CoV-2 in collaboration with Cellspect Co., Ltd.
Original publication
Funakoshi et al.: Response to mRNA SARS-CoV-2 vaccination evaluated by B-cell receptor repertoire after tixagevimab/cilgavimab administration. British Journal of Haematology(2023). DOI: doi.org/10.1111/bjh.18932.