To make advances in using microbes to sustainably produce materials, it is necessary to find new molecular tools, or “enzymes” — but this is labor intensive. A Kobe University team now developed a technique that can classify thousands of candidates and a workflow that can evaluate representatives overnight, in what may become a fundamental technology for biomanufacturing.
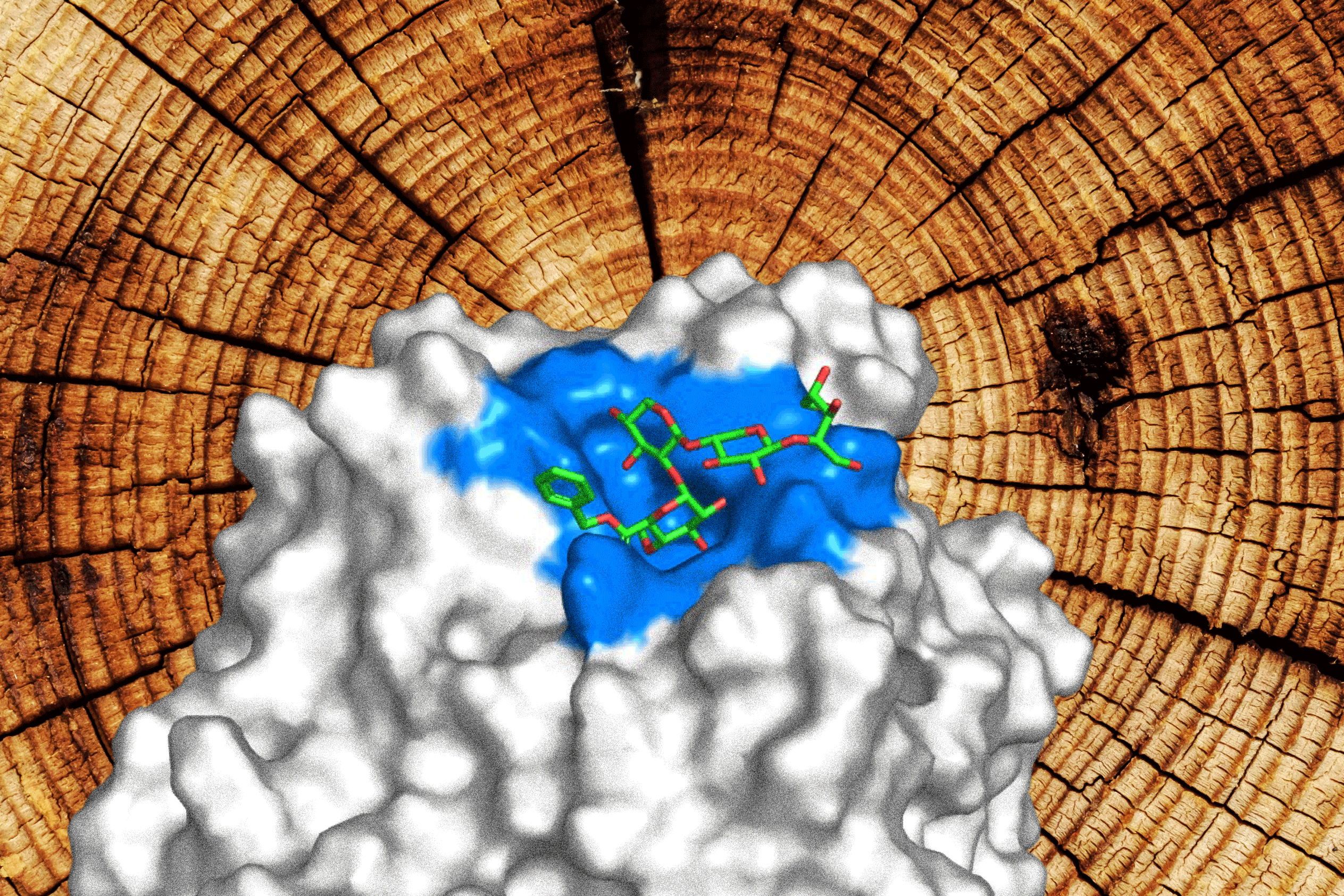
HASUNUMA Tomohisa and his team developed a workflow that allows them to screen a large variety of enzymes for a given function and tried it on a class of almost 7,000 enzymes that are involved in a process needed to produce the raw materials for fuels, plastics and flavors. In the journal ACS Catalysis, the Kobe University team now reports that this approach allowed them to identify an enzyme that has a productivity up to 10 times higher than that of the current industry standard. © Kobe University (CC BY)
As oil reserves dwindle and prices soar, microorganisms can help us produce useful chemicals and fuels from renewable resources. They can convert raw materials into products under mild conditions through the use of specialized molecular tools called “enzymes.” Finding appropriate enzymes, modifying them and putting them together into molecular assembly lines is what “biomanufacturing” is all about. Kobe University bioengineer HASUNUMA Tomohisa says: “Who controls enzymes controls biomanufacturing. There are easily accessible databases with more than 200 million enzyme entries, but much of the information on them is speculative and it’s time consuming and labor intensive to confirm their function.”
To solve this issue, Hasunuma and his team came up with a new way of automatically grouping large numbers of enzymes in a way that makes it easy to select a set of meaningful representatives and focus research on those. In addition, they developed a robotic system that can test the activity of the representative enzymes on a range of raw materials within one day. Together, this would allow them to screen a large variety of enzymes for a given function, and they decided to try it on a class of almost 7,000 enzymes that are involved in a process needed to produce the raw materials for fuels, plastics and flavors.
In the journal ACS Catalysis, the Kobe University team now reports that this approach allowed them to identify an enzyme that has a productivity up to 10 times higher than that of the current industry standard. What’s equally important, though, is that the newly identified enzyme is also as versatile as that standard; that is, it can perform the reaction on a broad range of raw materials. “Most of all, this finding demonstrates that our approach is able to identify hitherto unrecognized, highly active and versatile enzymes from these databases,” Hasunuma sums up the achievement.
The bioengineer, however, is also keen to point out another benefit of their method, saying: “The large amount of data on both the differences between the enzymes and the differences in their versatility allows us to pinpoint which parts of the enzyme are probably responsible for a given desirable trait. This not only helps us to clarify the action of an enzyme and improve that function in a more targeted way, but also lets us search for that structure in yet other enzymes.”
Hasunuma hopes that the technology his team developed will be so useful that it becomes a fundamental technology for biomanufacturing just like the databases themselves. But he is already looking for the next thing, explaining: “Our technology lets us connect enzyme structure with function on a large scale — this is perfect training material for an AI. We are thinking about developing an AI that can then turn around and use the data in the databases to predict the function of the enzymes more accurately.”
Acknowledgements
This research was funded by the New Energy and Industrial Technology Development Organization (grants P16009, P20011) and the Japan Society for the Promotion of Science (under the “Program for forming Japan’s peak research universities (J-PEAKS)”). It was conducted in collaboration with a researcher from the Tokyo University of Agriculture and Technology.
Original publication
R. Hidese et al.: Identification of sub-family-specific residues within highly active and promiscuous alcohol dehydrogenases. ACS Catalysis (2025). DOI: 10.1021/acscatal.5c02764
Release on EurekAlert!
Finding the enzymatic needle in the database haystack
Inquiries
For inquiries, please contact gnrl-intl-press[@]office.kobe-u.ac.jp