An international research team has clarified the regulatory mechanism of the ubiquitin-proteasome system (*1) in recognizing and repairing DNA that has been damaged by ultraviolet (UV) light. The investigators at Kobe University (Japan), the National Institute of Health Sciences (Japan), the Catholic University of Louvain (Belgium), Kyoto University (Japan), and the National Institute of Genetics (Japan) have published their results in the journal Scientific Reports.
Main points
- The researchers developed a microscope system whereby they could induce DNA damage to any intracellular location via UV light stimulation and observe cellular responses in real time.
- They observed that proteasome, which is a multi-protein complex involved in protein degradation, quickly accumulated at the sites of the UV light-induced DNA damage.
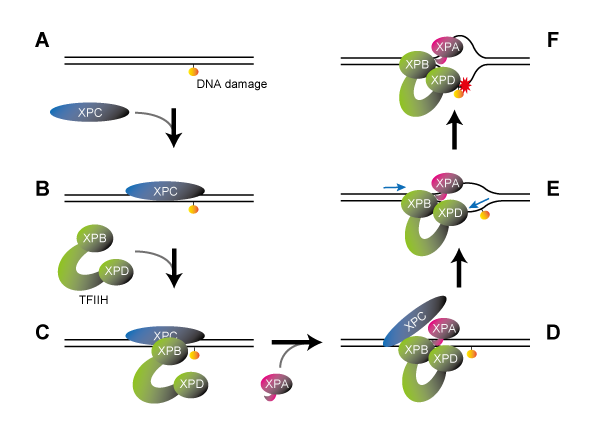
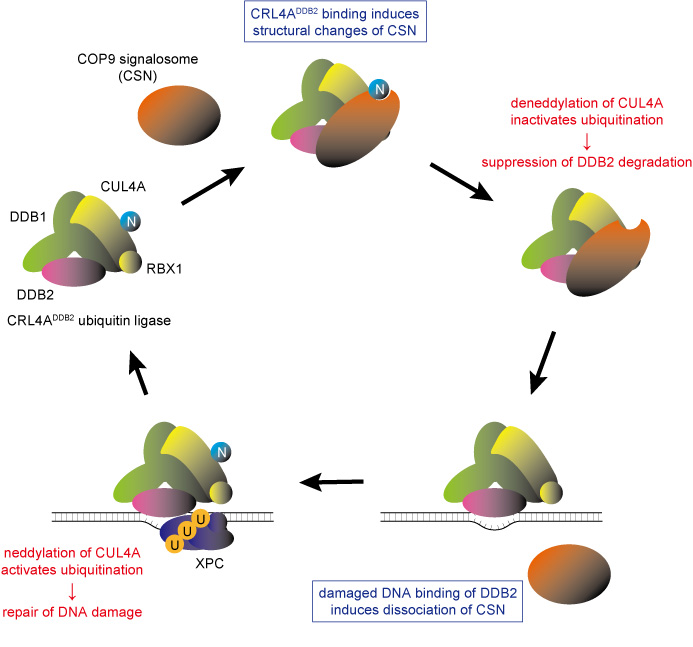
- This research has illuminated that the enzymatic activity and structural integrity of the proteasome are vital for the DNA damage recognition pathway, which is mediated by DDB2 proteins. Mutation in the DDB2 gene occurs in the genetic disorder xeroderma pigmentosum (*2).
Research Background
UV light from the sun is very harmful to living organisms because it can damage their genes. This is called DNA damage and normally it is fixed by the repair system in our cells, preventing us from experiencing adverse effects resulting from exposure to sunlight during the course of our daily lives. However, patients with xeroderma pigmentosum (XP) are born with malformations in this repair system, which means that their bodies are unable to sufficiently repair DNA damage caused by UV light. Consequently, they are predisposed to developing skin cancer in sun-exposed areas.
Intracellular proteins are generated and degraded as required, and the ubiquitin-proteasome system is known to play an important role in managing this degradation process. It has been previously shown that the ubiquitin-proteasome system coordinates cellular responses to repair the UV-induced DNA damage. However, the detailed mechanism behind this had not been clarified until now.
Research Findings
The international team, led by Prof. SUGASAWA Kaoru at the Biosignal Research Center, Kobe University, developed a custom microscope system (*3) which allowed them to successfully observe the dynamic behaviors of various intracellular proteins in response to UV-induced DNA damage (Figure 1, left).
The DDB2 protein is one of the gene products responsible for XP and is important for recognizing UV-induced DNA damage. Utilizing the custom microscope system enabled the researchers to make a new discovery: they found that the DDB2 protein works together with the ubiquitin-proteasome system to promote DNA repair. First of all, the multi-protein complexes, proteasomes, quickly accumulated at DNA damage sites depending on the presence of DDB2 proteins. This suggests that the proteasome’s protein degradation function could be activated following the damage recognition and repair (Figure 1, right).
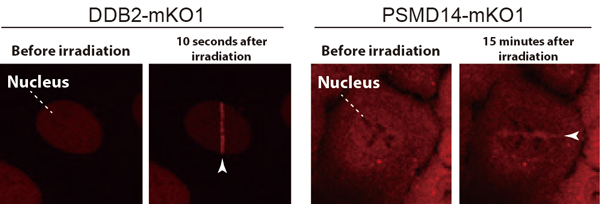
The white arrows indicate the molecules with attached fluorescent proteins that have accumulated at UV-induced DNA damage sites.
Furthermore, using an inhibitor to suppress this proteasome activity caused the proteasome to accumulate in a specific region of the nucleus, trapping the DDB2 protein and making it unable to participate in DNA damage repair (Figure 2). On the other hand, suppressing the expression of proteasome subunits compromised proper assembly of the proteasomes and the aforementioned proteasome aggregation was no longer observable. However, the absence of proteasomes severely suppressed the accumulation of DDB2 proteins at DNA damage sites.
These results revealed for the first time that proteasomes’ protein degradation activity and architectural integrity are involved in the regulation of DDB2 protein-mediated DNA damage repair via separate mechanisms.
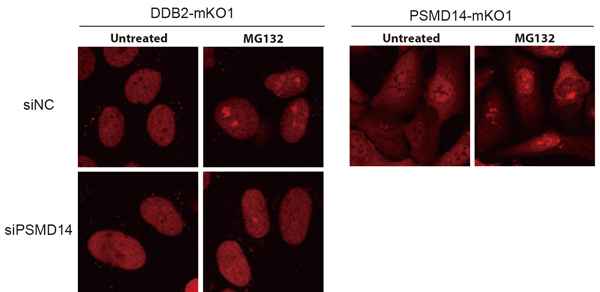
Inhibiting proteasome with MG132 causes both DDB2 and proteasome to accumulate in the vicinity of the nucleoli. However, this aggregation can be alleviated by suppressing PSMD14 expression.
Further Developments
The results of this study show that the action of the DNA damage recognition mechanism is essential for enabling DNA damage to be efficiently repaired. Furthermore, this understanding will also contribute towards clarifying the onset mechanisms of diseases such as skin cancer, in addition to the development of treatments to suppress this onset.
Glossary
- *1 Ubiquitin-proteasome system
- In this system, the small protein molecule, ubiquitin, is used as a ‘molecular marker’ for unnecessary protein. A large multi-protein complex, proteasome, recognizes and degrades the ubiquitin-marked proteins. This is one of the important systems for protein quality control in the cell.
- *2 Xeroderma pigmentosum (XP)
- A rare autosomal recessive genetic disorder that is known to occur more frequently in Japanese people. It is caused by malfunctions in the nucleotide excision repair system that repairs damage caused to DNA by UV light (e.g., sunlight).
- *3 Custom microscope system
- A system that enables the cell response to UV-induced DNA damage to be observed in real time, with a confocal laser scanning microscope as the main component. The system was equipped with a 780 nm femtosecond fiber laser. According to the principle of three-photon absorption, this laser allowed the researchers to apply stimulation to the cells that was comparable to stimulation by 260 nm UV light.
Acknowledgements
This research was funded by a Grant-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (JSPS) (Grant number: JP16H06307).
Journal Information
- Title
- “Functional impacts of the ubiquitin–proteasome system on DNA damage recognition in global genome nucleotide excision repair”
- DOI
- 10.1038/s41598-020-76898-2
- Authors
- Wataru Sakai, Mayumi Yuasa-Sunagawa, Masayuki Kusakabe, Aiko Kishimoto, Takeshi Matsui, Yuki Kaneko, Jun-ichi Akagi, Nicolas Huyghe, Masae Ikura, Tsuyoshi Ikura, Fumio Hanaoka, Masayuki Yokoi, Kaoru Sugasawa
- Journal
- Scientific Reports