A research group consisting of Associate Professor FUKAYAMA Hiroshi (Kobe University, Graduate School of Agricultural Science) and Professor MATSUMURA Hiroyoshi (Ritsumeikan University) et al. have succeeded in greatly increasing the catalytic activity of Rubisco (*1), the enzyme which fixes carbon from CO2 in plant photosynthesis. The research team also hypothesized the mechanism which determines the catalytic activity of Rubisco, based on structural analysis of the proteins.
In the future, it is hoped that increasing the photosynthetic ability of agricultural crops will lead to increased yields.
These results were published in the international scientific journal Molecular Plant on August 31.
Main points
- Photosynthesis determines a plant’s growth rate. The low activity of the enzyme Rubisco, which is the catalyst for the reaction that turns CO2 into organic carbon, limits the rate of photosynthesis.
- Rubisco consists of two types of protein; large subunits (RbcL) and small subunits (RbcS). RbcS is an important factor for determining the speed of the catalyst.
- A hybrid Rubisco consisting of rice RbcL and sorghum RbcS demonstrated a catalytic rate that was approximately 2 times higher than that of rice Rubisco. This is believed to be the first time in the world that such a large increase in Rubisco activity has been achieved.
- The 102 amino acid found inside RbcS is isoleucine in rice and leucine in sorghum. Analysis of the protein structures indicated the possibility that the difference in amino acid type could affect the catalytic activity.
- It is hoped that the method to improve photosynthesis demonstrated in this study could be applied to many other crops which, like rice, have low Rubisco activity such as wheat, soybean and potato.
Research Background
Growth speed in plants is mainly determined by photosynthetic ability. Thus improving photosynthesis in agricultural crops can increase their yield. In photosynthesis, Rubisco is an enzyme that acts as the initial catalyst for the reaction which turns CO2 into organic carbon. However, Rubisco has two major drawbacks which limit photosynthesis: its catalytic activity is very low, and it can be inhibited by O2 (ie. Rubisco can mistakenly fix to O2 molecules instead of CO2 molecules, creating a toxic compound that needs to be recycled by the plant).
Rubisco’s catalytic activity varies depending on the type of plant. Most major crops, such as rice, wheat and soybean are C3 plants that use regular photosynthesis. C4 plants, such as corn and sugarcane, on the other hand, have acquired a mechanism to concentrate CO2 (the C4 photosynthetic pathway).
The catalytic rate is low in C3 plants, whereas in C4 plants it tends to be high. Rubisco with high catalytic activity tends to be inhibited easily by oxygen, therefore it cannot function effectively in atmospheric conditions where there is a low concentration of CO2 if the plant doesn’t have a CO2-concentrating mechanism. However, as the amount of atmospheric CO2 is continuing to increase, it is believed that if C3 plants had the same highly active type of Rubisco as C4 plants then this could be utilized to improve photosynthetic ability.
Research Findings
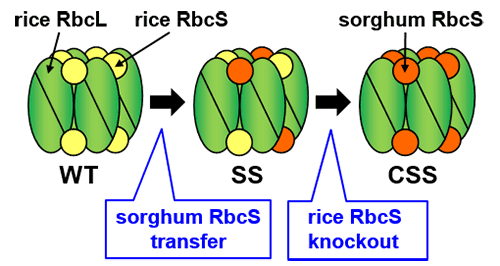
WT (Wild Type, unmodified rice), SS (rice with sorghum RbcS), CSS (rice with sorghum RbcS transferred/rice RbcS knocked out).
Rubisco is made up of two types of protein- large subunits (RbcL) and small subunits (RbcS) (Figure 1). The sequence of the amino acids in RbcS varies greatly between species. This team has been focusing on conducting research into RbcS. They genetically modified rice (a C3 plant) by transferring RbcS from the C4 plant sorghum, successfully increasing the catalytic rate of rice Rubisco 1.5 times. This rice with sorghum RbcS inserted (SS line), produced a chimera form of Rubisco from both sorghum RbcS and rice RbcS. Next, the rice RbcS gene was knocked out in the sorghum RbcS incorporated rice plants using CRISPR/Cas9 gene editing.
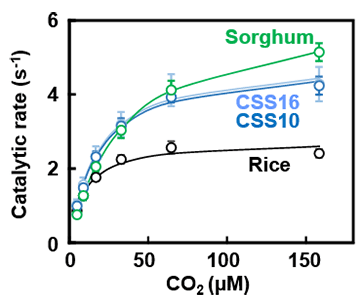
Graph showing the relationship between CO2 concentration and the catalytic activity of Rubisco. CSS10 and CSS16 are sorghum RbcS incorporated/rice RbcS knockout lines.
In this CSS line (sorghum RbcS transferred/rice RbcS knocked out), the rice RbcS was completely replaced by sorghum RbcS, producing hybrid Rubisco. This approximately doubled the catalytic rate to that which is equivalent to C4 plants (Figure 2). Although many researchers have been able to improve Rubisco’s catalytic characteristics, there have been no examples of such a large increase being achieved. Furthermore, CSS line plants demonstrated a higher photosynthetic rate than unmodified (wild type) rice under high CO2 conditions, even though the amount of Rubisco in their leaves was over 30% less.
Subsequently, the researchers conducted x-ray crystallography (*2) in order to illuminate the mechanism by which sorghum RbcS increases Rubisco’s catalytic activity. RbcL is present in Rubisco’s catalytic site. Near this catalytic site, there is a structure called RbcS βC (Figure 3). The 102 amino acid found in βC is isoleucine in rice and leucine in sorghum. Leucine has smaller molecules than isoleucine. Therefore, it is thought that in sorghum RbcS the gaps between amino acid molecules become bigger, making the reaction site more pliable and thus increasing catalytic activity. Although further research is necessary to prove this, it is believed to be a previously unproposed ground-breaking theory for Rubisco research.
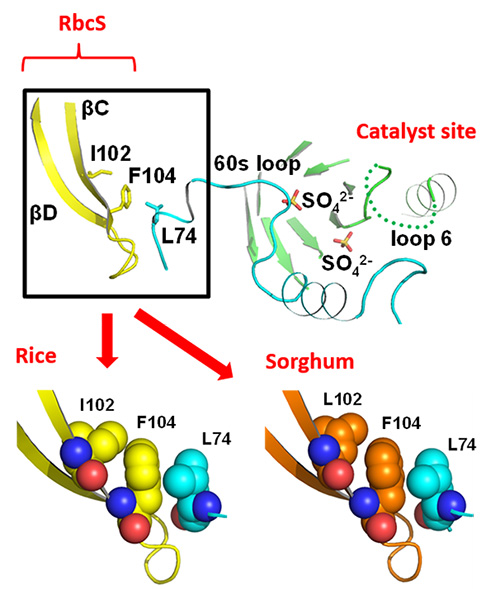
The 102 amino acid is isoleucine (I102) in rice RbcS and leucine (L102) in sorghum RbcS. The gap between L102 and the neighboring phenylalanine (F104) is larger than the gap between I102 and F104.
Conclusion
The CSS line produced in this study demonstrated high photosynthetic ability, however crop yield was not improved. Hopefully, it will be possible to vastly improve plant growth and productivity through appropriate control of Rubisco levels.
The current research used the C3 plant rice, however it is vital to consider the applications of this methodology and investigate whether or not the same strategy can be used to increase Rubisco’s catalytic activity in other major crops, such as wheat, soybean and potato.
It is thought that the 102 amino acid is an important determinant of the catalytic activity. Further research is being carried out to investigate this; for example by replacing only the amino acid at the 102 site with another amino acid and producing Rubisco.
Glossary
- *1 Rubisco (Ribulose-1,5-biphosphate carboxylase/oxygenase)
- In photosynthesis, the enzyme Rubisco is the catalyst for the initial reaction that fixes CO2. It is not only the catalyst for the photosynthesis reaction (carboxylase reaction) but also serves as the catalyst for the oxygenase reaction where O2 is the substrate.
Plants accumulate large amounts of Rubisco in their leaves to compensate for the very slow rate of the catalyst. Around half of the soluble proteins in rice plant leaves are Rubisco and it is known to be the most prevalent protein on Earth. - *2 X-ray crystallography
- A method to determine the spatial coordinates of the atoms that make up a crystal by analyzing the diffraction obtained from x-raying a single crystal. This is one way of determining the structural arrangement of proteins. It is necessary to produce protein crystals.
Acknowledgements
This study received the following research funding:
- JSPS KAKENHI (17H05732, 18K06094, 19H04735, 19K07582, 24580021, 15H04443)
- Ritsumeikan University (R-GIRO)
- Osaka University (CR-18-05 and CR-19-05)
- Platform Project for Supporting Drug Discovery and Life Science Research from AMED (JP19am0101070)
Journal Information
- Title
- “Hybrid Rubisco with complete replacement of rice Rubisco small subunits by sorghum counterparts confers C4-plant-like high catalytic activity”
- DOI
- 10.1016/j.molp.2020.08.012
- Authors
- Hiroyoshi Matsumura1,*, Keita Shiomi2, Akito Yamamoto2, Yuri Taketani3, Noriyuki Kobayashi2, Takuya Yoshizawa1, Shun-ichi Tanaka1, Hiroki Yoshikawa1, Masaki Endo4, Hiroshi Fukayama2,*
1 College of Life Sciences, Ritsumeikan University
2 Graduate School of Agricultural Science, Kobe University.
3 Faculty of Agriculture, Kobe University
4 National Agriculture and Food Research Organization
* Corresponding author - Journal
- Molecular Plant