A research collaboration has discovered a new mechanism by which glucose that was captured by E. Coli is then secreted from the bacterial cell as glucose-6-phosphate (G6P). This finding has significant applications for improving methods to sustainably produce useful aromatic compounds and raw materials for the medicine, food and chemical industries (among others) from biomass. The research group consisted of Associate Professor TANAKA Tsutomu of Kobe University’s Graduate School of Engineering, Special Postdoctoral Researcher FUJIWARA Ryosuke and Researcher NODA Shuhei et al. of RIKEN Center for Sustainable Resource Science, and Professor UMETSU Mitsuo et al. of Tohoku University’s Graduate School of Engineering.
Based on the discovered mechanism, the researchers successfully developed a new technique to increase the production of the target compound by combining metabolic engineering and cell surface engineering.
It is hoped that by applying this technique, it will be possible to greatly increase the amount of target compound produced by adding a small amount of ‘metabolic spice’ to the microbe’s culture broth.
These research results were published in Metabolic Engineering on March 5, 2022.
Main points
- The researchers discovered that a portion of the glucose captured by E. Coli was secreted as glucose-6-phosphate (G6P).
- They successfully developed a novel technique that can increase the production of various compounds by trapping the secreted G6P on the surface of the bacteria.
- It is hoped that the new technique can be applied to ‘metabolic spices’. This will enable large amounts of useful compounds to be produced by simply adding a small amount of ‘metabolic spice’ to the microbe’s culture broth.
Research Background
A big advantage of products derived from biomass (low-cost renewable resources that are abundant in nature, such as grass and trees) is that they are carbon neutral (*1); their production and disposal do not increase atmospheric CO2 levels. In bioproduction technologies (*2), this biomass is utilized as a raw material and microbes are applied to it to produce target compounds (for example, aromatic compounds for chemical, pharmaceutical and food industries, among others). The development of these technologies contributes towards SDGs and is vital for realizing a low carbon society.
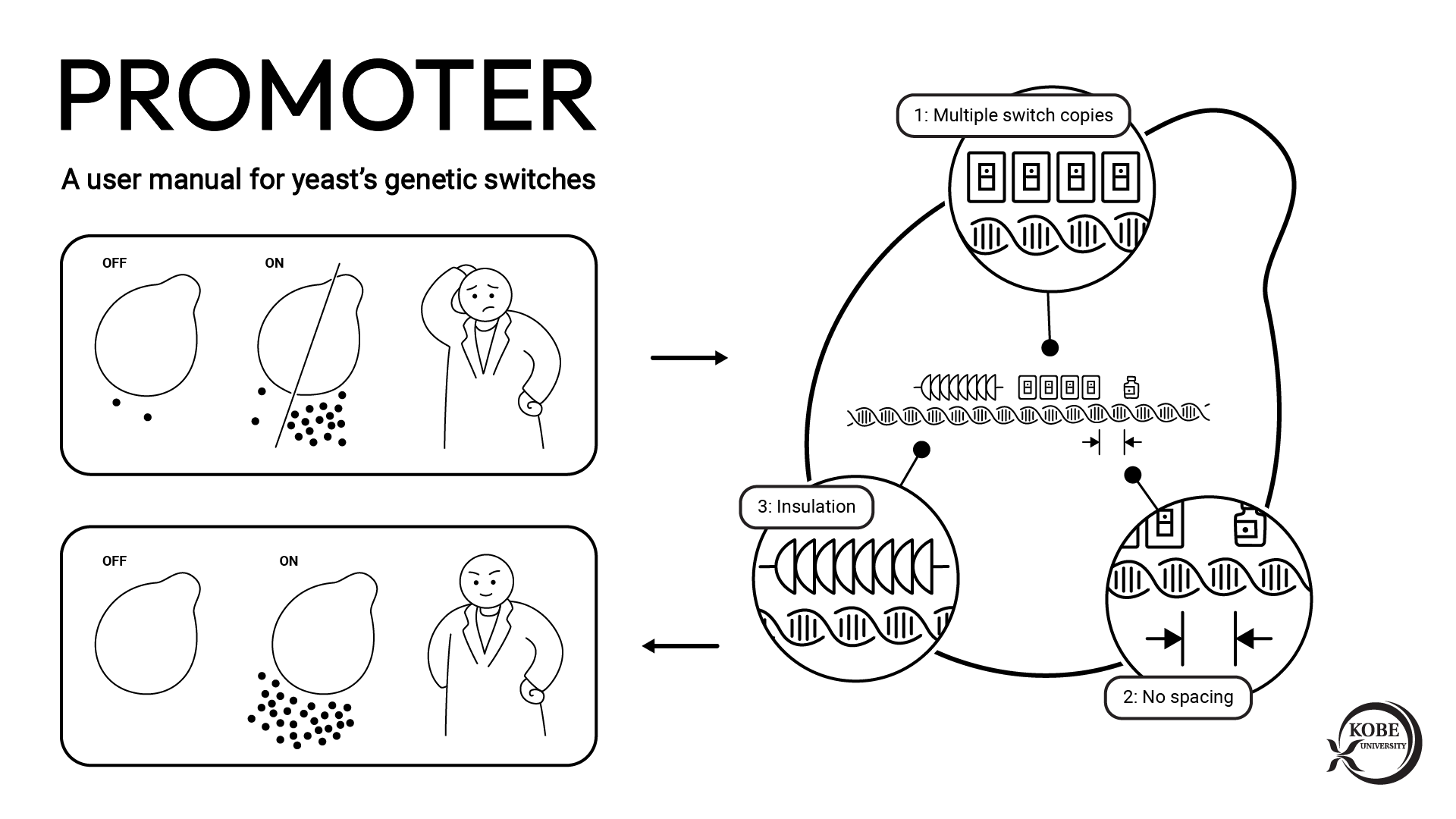
Associate Professor Tanaka’s research group have exhibited biomass decomposition enzymes on the surface of a variety of microbes, and have developed cell surface engineering technology to improve the spatial resolution of biomass. Microbes degrade the sugars found in plant biomass such as cellulose and cellooligosaccharide into glucose. In their previous study, the researchers metabolically engineered the microbes so that the degraded glucose was captured by the microbes and used solely to produce the target compound.
In the course of this research, they discovered a new phenomenon completely unrelated to biomass degradation. They found that production of the target compound was increased by enzyme surface expression. Utilizing this phenomenon, they demonstrated that it was possible to increase the rate and amount of biomass-derived compound production, contributing towards the decarbonization of matter production. The group then conducted further research with the aims of understanding the mechanism behind this phenomenon and applying it to metabolic spices.
Research Methodology
This research group discovered a completely new phenomenon whereby a portion of the glucose captured by the model microorganism E. coli is ejected from the bacterial cell as glucose-6-phosphate (G6P). Normally, microbes continuously convert glucose (their source of nutrition) into G6P and capture it inside their cells. Until now, it was thought that G6P wasn’t ejected from the bacterial cells once captured. This study revealed a novel mechanism by which E. coli ejects G6P, contradicting the commonly accepted theory. The biomass degradation enzyme is expressed on the cell surface of the microbe and ejects the G6P. The researchers found that temporarily trapping this G6P stimulated the metabolism inside the E. coli cells thus increasing the production of the target compound (Figure 1).
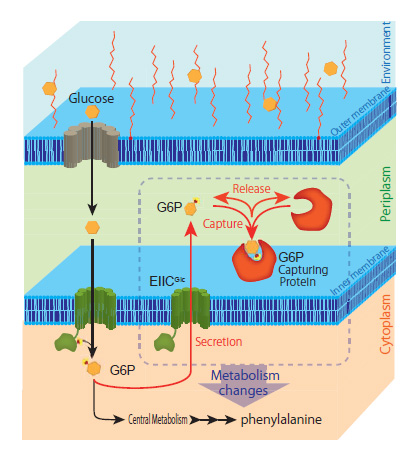
Next, the research group utilized this mechanism to localize various proteins that can trap G6P on the bacterial cell surface. In this way, they developed a new technique to increase target compound production. They successfully demonstrated the novel technique by producing increased amounts of the aromatic amino acid phenylalanine (Figure 2).
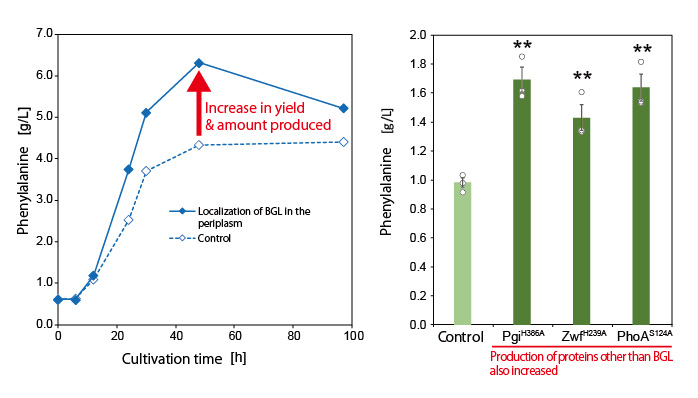
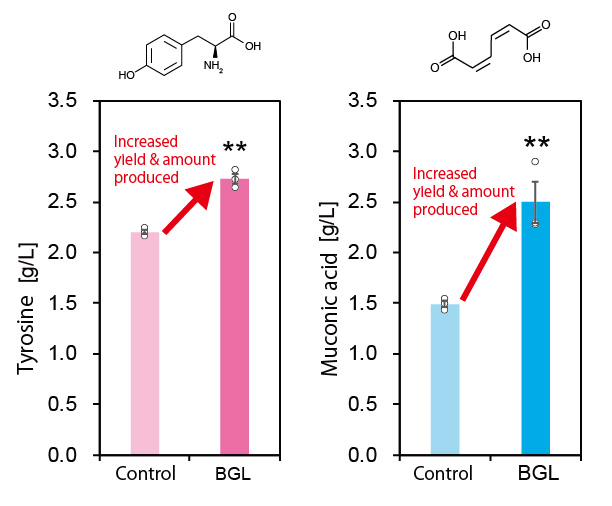
Furthermore, they also increased the production of the aromatic amino acid tyrosine and muconic acid (a useful Dicarboxylic acid) by using the developed technique (Figure 3). Muconic acid, in particular, is a very important and useful industrial chemical. It can be easily converted into adipic acid, an ingredient in nylon production and is also used as a raw material in the production of various drugs and chemical products. These results demonstrate that this new technique is highly versatile and can be applied to the production of various chemical compounds.
Further Developments
The research group has confirmed that, as an alternative to expressing proteins on the surface of bacterial cells, it is possible to make the same phenomenon occur by adding a special molecule to the culture broth. The researchers have termed small molecules possessing this characteristic ‘metabolic spices’ and are currently working on developing them (Figure 4). They hope to increase target compound production merely by adding a small amount of metabolic spice to the microbe’s culture broth, without the need to genetically engineer the microbe.
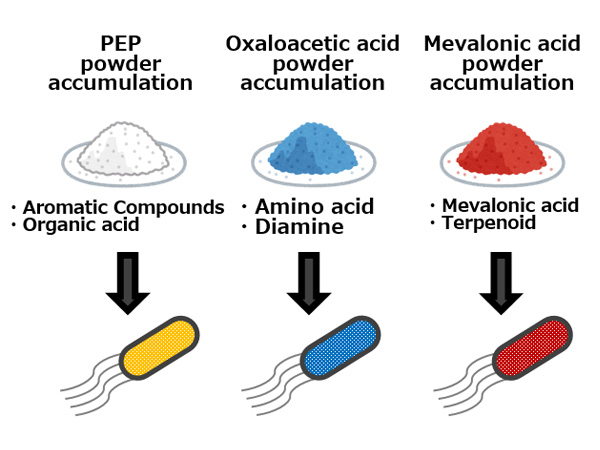
By adding a small amount of a ‘metabolic spice’ to the culture broth, target compound production can be significantly increased.
In addition, this technique enhances specific metabolic pathways and increases the supply of precursors to various useful substances. It can also be applied even in cases where the final product cannot be determined. Furthermore, the results of this study have shown that a moderate level of selectivity and affinity with G6P is sufficient. The metabolic spice approach is different to existing approaches for discovering chemicals with high selectivity and affinity. Therefore, it is likely that there are new candidates for the metabolic spice approach among the ‘rejected chemicals’ catalogued in the chemical libraries kept by various industries. It is hoped that this will lead to new academia-industry collaborations even with companies that have no prior involvement in bioproduction.
Glossary
- 1. Carbon Neutral
- A concept to keep the amount of atmospheric CO2 constant by offsetting carbon emissions with carbon removal, or by eliminating carbon emissions altogether. Using biomass instead of fossil fuels is an example of a carbon neutral process.
- 2. Bioproduction technologies
- Technologies that utilize renewable biomass resources as a raw material from which to produce biofuels, bioplastic and pharmaceutical ingredients among others.
Acknowledgements
This research received the following funding:
- The Japan Science and Technology Agency (JST)’s Mirai program: Small start-type (feasibility study) “Realization of a Low Carbon Society, a global issue”: Realization of a low carbon society through game changing technologies. Research title: ‘Creation of PEP-accumulating chassis strains by cell surface engineering and metabolic engineering’ (Grant No. JPMJMI17EI).
- Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (B) (Grant Number 19H02526).
- Special Postdoctoral Researcher Program, RIKEN Center for Sustainable Resource Science.
- Grant-in-Aid for JSPS Fellows.
Journal Information
- Title
- “G6P-capturing molecules in the periplasm of Escherichia coli accelerate the shikimate pathway”
- DOI
- 10.1016/j.ymben.2022.03.002
- Authors
- Ryosuke Fujiwara, Mariko Nakano, Yuuki Hirata, Chisako Otomo, Daisuke Nonaka, Sakiya Kawada, Hikaru Nakazawa, Mitsuo Umetsu, Tomokazu Shirai, Shuhei Noda,* Tsutomu Tanaka,*, Akihiko Kondo
*Corresponding Author - Journal
- Metabolic Engineering